- Cart 0
- English
RCR, RT-PCR, qPCR, RT-qRCR Learned?
August 18, 2025
Clicks:119
Polymerase Chain Reaction (PCR) was developed by Dr. Kary Mullis in the 1980s. The technology has been likened to a "molecular copier" for its ability to identify specific DNA sequences and synthesize large copies quickly and accurately. Various derivative techniques based on the original PCR method are currently developed. Real-time PCR, also known as quantitative PCR (qPCR), combines PCR amplification and detection in one step. Another technique called reverse transcription polymerase chain reaction (RT-PCR) uses RNA as a nucleic acid initiation template.
★PCR : Repeated heating and cooling cycles of a reaction mixture containing DNA template, DNA polymerase, primers, and dNTPs. [1] Each cycle roughly doubles the amount of DNA, because in the next cycle, a new strand of DNA becomes the template for replication. This leads to an exponential increase in DNA quantities.
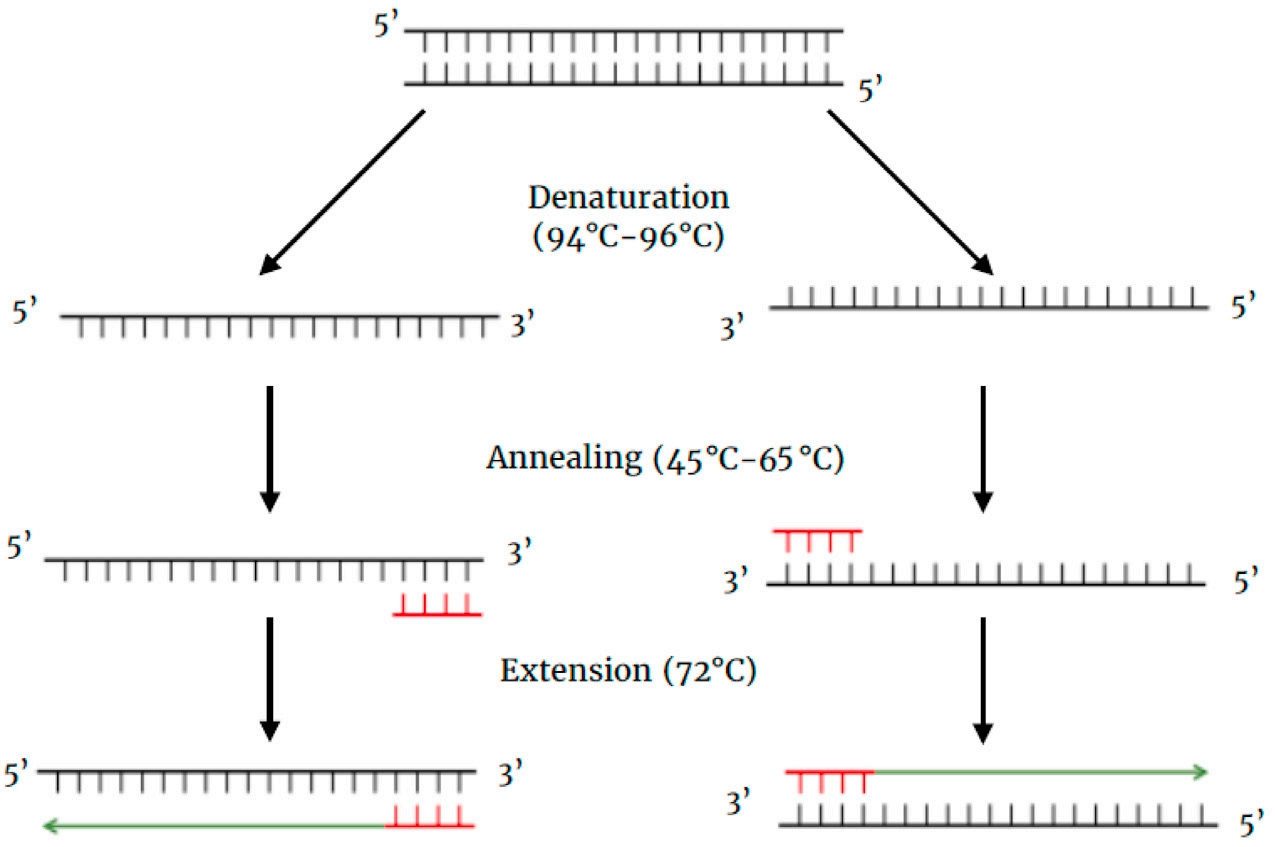
Figure 1 First cycle of PCR [1]
★RT-PCR (Reverse transcription PCR): Reverse transcription PCR can use RNA as a template to generate complementary DNA (cDNA). A single-stranded copy of cDNA can be generated using reverse transcriptase. This can be done using oligomeric (dT) primers that bind to the PolyA tail of the RNA, or using random hexamers (primers six to nine bases long that bind at multiple points of the RNA transcript). In general, a mixture of the two primers is considered the best because of its ability to amplify PolyA tail RNA (mainly mRNA) and PolyA-free RNA (tRNA, rRNA, etc.). It is then amplified with DNA polymerase to generate double-stranded cDNA, which is sent to a standard PCR-based amplification process.
★qPCR (Quantitative PCR): It is a specified amount of real-time PCR, that is, DNA is PCR amplified in real time and measured by fluorescent probes (most commonly intercalation dyes or hydrolysis probes) to quantify the PCR products. Used to detect the presence of pathogens and to determine the copy number of associated DNA sequences. SYBR Green I is the most commonly used DNA-binding fluorescent dye. When bound to double-stranded DNA, it emits fluorescence, and the fluorescence intensity increases proportionally to the concentration of PCR products.
The advantage of quantitative PCR over standard PCR is that it is possible to observe which reactions are working in real time without the need for an agarose gel. It also enables real quantitative analysis.
★RT-qPCR (Reverse Transcription Quantitative real-time PCR): This is a technique that combines RT-PCR with qPCR for Reverse Transcription Quantitative real-time PCR. Changes in gene expression were rapidly detected by measuring RNA levels using cDNA in qPCR reactions.
RT-qPCR can be used to study changes in gene expression when a model system is treated with inhibitors, stimulants, small interfering RNAs (siRNAs), or gene knockout models, etc. This technique is also frequently used to detect changes in expression before (as quality control) and after (confirmation changes) RNA-Seq experiments.
The final step in RT-qPCR sample preparation is the generation of cDNA. In addition to considering primers, the generation of cDNA can be part of a qPCR experiment (called one-step RT-qPCR) or separately from qPCR (two-step RT-qPCR) (Fig. 3).
|
Method |
One-step RT-qPCR |
Two-step RT-qPCR |
|
advantage |
Less experimental variation, less risk of contamination, and enables high-throughput screening |
More reactions per sample and flexible priming options |
|
shortcoming |
Limited number of samples used |
More optimization is required |
|
Applications |
Clinical screening |
Large-scale gene expression analysis |
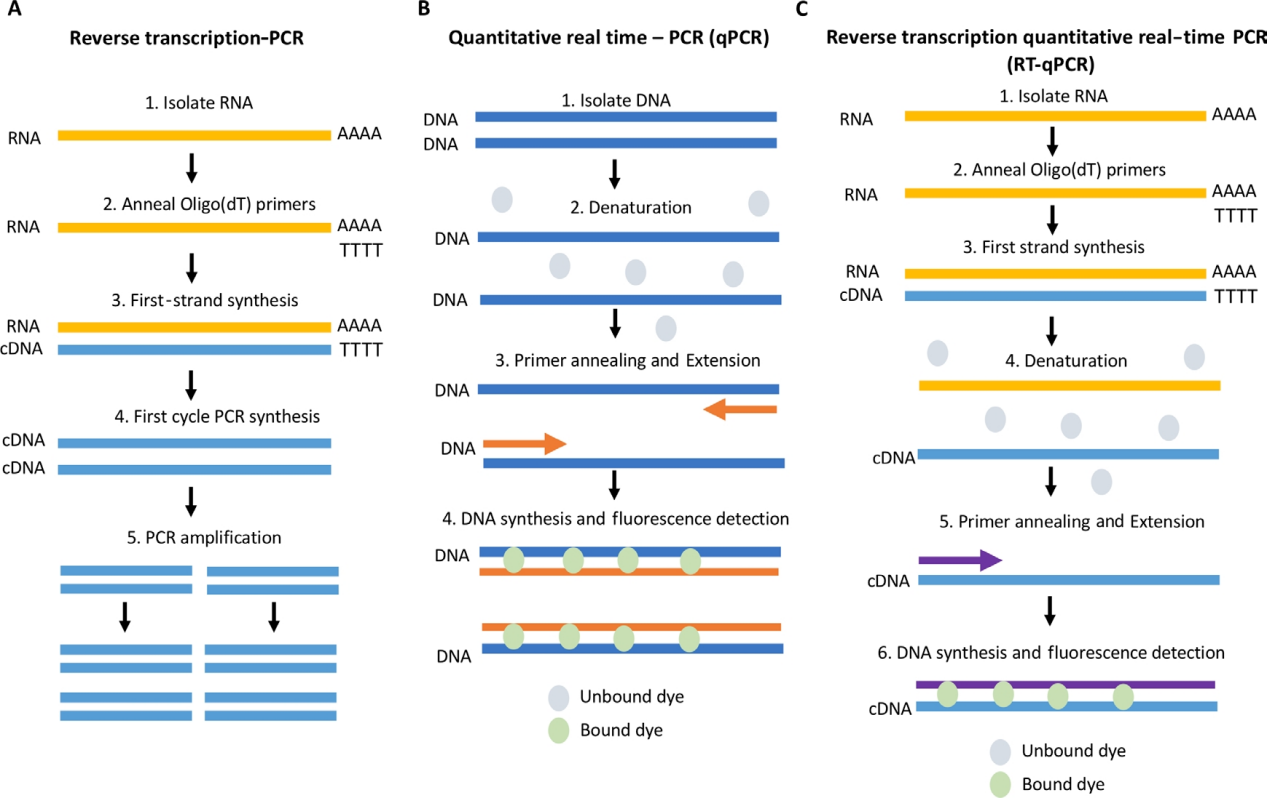
Figure 2 Schematic of RT-PCR, qPCR and RT-qPCR [2]
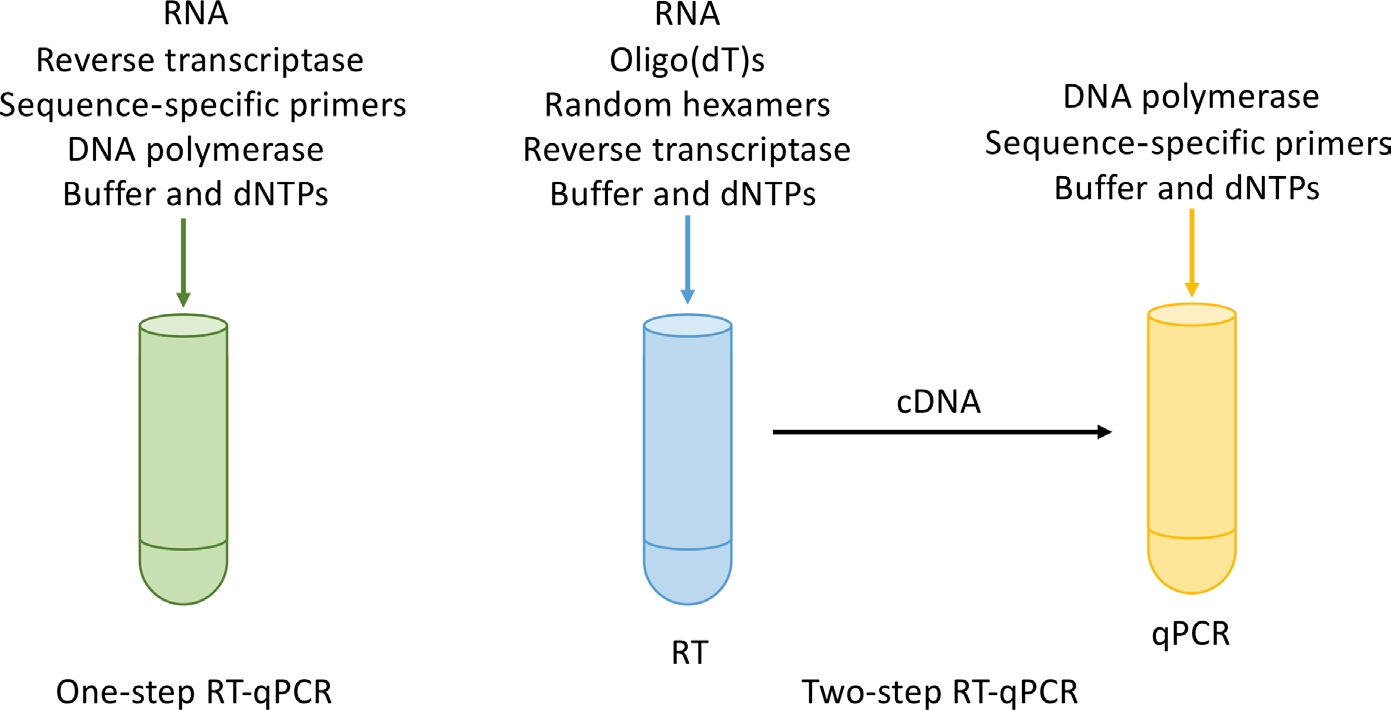
Figure 3 One-step RT-qPC and two-step RT-qPCR [2]
★ddPCR (Droplet-based Digital PCR): The sample passes through the microfluidic chip to form a segmented flow, in which the sample is divided into tens of thousands of droplets can act as a PCR reaction chamber, and these droplets are separated by immiscible liquids (such as mineral oil) to form an emulsion [3]. The emulsions were collected in vials, PCR was performed, and the samples were subsequently processed by flow cytometry to count the number of PCR-positive droplets. Or the emulsion is fed into a plastic chip to form a single layer of droplets, thermal cycling is performed and fluorescence images of the droplets are captured and evaluated [4]. Compared with qPCR, ddPCR has the advantage of higher accuracy and lower absolute quantitative coefficient of variation [5]. PCR multiplexing is typically performed by probe-based fluorescence, with each DNA/RNA having a different excitation color. Also used to generate libraries for single cell RNA sequencing.

Fig. 4 Schematic of ddPCR [6]
References:
[1] Jalali M, Zaborowska J, Jalali M. The polymerase chain reaction: PCR, qPCR, and RT-PCR[M]//Basic science methods for clinical researchers. Academic Press, 2017: 1-18.
[2] Zhang H, Tang K, Wang B, Duan CG, Lang Z, Zhu JK. Protocol: a beginner's guide to the analysis of RNA-directed DNA methylation in plants. Plant Methods. 2014 Jun 14;10:18. doi: 10.1186/1746-4811-10-18. PMID: 24955108; PMCID: PMC4065543.
[3] Schiffman M, Bauer H, Lorincz A et al. Comparison of Southern blot hybridization and polymerase chain reaction methods for the detection of human papillomavirus DNA. J. Clin. Microbiol.29(3), 573–577 (1991).
[4] Madic J, Zocevic A, Senlis V et al. Three-color crystal digital PCR. Biomol. Detect. Quant. 10, 34–46 (2016).
[5] Hindson CM, Chevillet JR, Briggs HA et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10, 1003 (2013).
[6] Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018 Aug 7;50(8):1-14. doi: 10.1038/s12276-018-0071-8. Erratum in: Exp Mol Med. 2021 May;53(5):1005. doi: 10.1038/s12276-021-00615-w. PMID: 30089861; PMCID: PMC6082860.
|
Item number |
Name |
Specifications |
|
2×Taq PCR Mix |
1mL×5 |
|
|
2×Taq PCR Master Mix(for PAGE) |
1mL×5 |
|
|
2× Xerox PCR Master Mix |
1mL×5 |
|
|
2×HotStart Taq plus Master Mix(Quick Load) |
1mL×5 |
|
|
2×Pfu Master Mix |
1mL×5 |
|
|
2×Long Taq PCR Master Mix |
1mL×5 |
|
|
Whole Genome Amplification Kit |
50T |
|
|
Reverse transcription one-tube three-generation premix |
100T |
|
|
First-strand cDNA Synthesis Mix With gDNA Remover |
100T |
|
|
One-Step RT-PCR Kit |
200T |
|
|
miRNA probe reverse transcription kit |
40T |
|
|
One-tube three-generation premix of degenomization and reverse transcription |
100T |
|
|
miRNA tailing reverse transcription kit |
40T |
|
|
Plasma miRNA tailing reverse transcription kit |
50T |
|
|
miRNA stem-loop reverse transcription kit |
50T |
|
|
RNase inhibitor |
1mL |
|
|
Two-Step RT-PCR Kit |
1kit |
|
|
2 × premixed real-time fluorescence quantitative rapid PCR reaction system |
1.7mL×3 |
|
|
SYBR One-Step qRT-PCR Kit |
100T |
|
|
Antibody Dye Method Quantitative PCR Premix (Universal ROX) |
1mL×5 |
|
|
2 × premixed real-time fluorescence quantitative rapid PCR reaction system (high concentration ROX) |
1.7mL×3 |
|
|
SYBR High-Sensitivity qPCR SuperMix |
1mL×5 |
|
|
miRNA stem-loop dye method fluorescence quantitative PCR kit |
200T |
|
|
2 × premixed real-time fluorescence quantitative rapid PCR reaction system (low concentration ROX) |
1.7mL×3 |
|
|
Fluorescence quantitative PCR reagent by miRNA probe method |
200T |
|
|
miRNA tailed dye method fluorescence quantitative PCR kit |
200T |
Absin provides antibodies, proteins, ELISA kits, cell culture, detection kits, and other research reagents. If you have any product needs, please contact us.
|
Absin Bioscience Inc. Email: worldwide@absin.cn |
 Follow us on Facebook: Absin Bio Follow us on Facebook: Absin Bio |
