- Cart 0
- English
【New Product Express: GeneScope】Explore the origins of fluorescence in situ hybridization
Fluorescence In Situ Hybridization (FISH) is a revolutionary advance in molecular biology and genetics. Not only does it provide us with a powerful tool to study the structure and function of genes, but it also helps us gain insight into the relationship between chromosomal abnormalities and disease. This article will take you through time to explore the origins of FISH technology and how it developed into an integral part of modern molecular biology.
1. The birth of in situ hybridization technology
The concept of In Situ Hybridization (ISH) technology can be traced back to the 60s of the 20th century. In 1969, scientists first reported in situ hybridization experiments using radiolabeled DNA probes, a technique that allowed researchers to locate specific DNA sequences in cell or tissue sections. Although this technique had some success at the time, its wide application was limited due to the safety and difficulty of handling radioisotopes.
2. Introduction of fluorescent labels
By the 70s of the 20th century, with the development of non-radioactive labeling technology, especially the emergence of labels such as biotin and digoxin, in situ hybridization technology began to become safer and easier to operate. However, the real turning point came in the 80s, when fluorescent labeling techniques were introduced into in situ hybridization, resulting in what we now know as fluorescence in situ hybridization (FISH). This advancement has dramatically improved the sensitivity and multiplexing of the assay, making it possible to detect multiple DNA or RNA sequences simultaneously on the same section.
3. Development of FISH technology
In the 90s, with the advancement of fluorescence microscopy technology and the diversification of fluorescent dyes, FISH technology ushered in a period of rapid development. Researchers began to use different colors of fluorescent dyes to label different probes to perform multiplex FISH experiments on the same section. This has led to a wide range of applications in the fields of chromosomal abnormality detection, cancer research, and gene expression analysis.
In the 21st century, with the development of genomics and personalized medicine, the application scope of FISH technology has been further expanded. Modern FISH technology is not only able to detect chromosome number and structural abnormalities, but also to pinpoint the location of genes on chromosomes, providing important information for the diagnosis and treatment of diseases. In addition, FISH technology has been used to monitor the cell cycle, study the dynamics of gene expression, and explore gene regulation mechanisms during cell differentiation and development.
4. Application of modern FISH technology
Fluorescence in situ hybridization technology has undergone decades of development and refinement from its initial concept to its widespread application. Not only has it greatly advanced our understanding of genes and chromosomes, but it has also provided a powerful tool for the diagnosis and treatment of diseases. As Product Manager for FISH Technology, we are proud to witness the growth of this technology and look forward to its greater role in future scientific research and clinical applications." As technology continues to advance, we believe that FISH technology will continue to unravel more chapters of life's mysteries for us.
5. Differences between different in situ hybridization techniques
Radioactive In Situ Hybridization (RISH), Digoxigenin In Situ Hybridization (DIG-ISH), and Fluorescence In Situ Hybridization, FISH) are all techniques for detecting and locating nucleic acid sequences, but they differ in markers, detection methods, and application areas. The following are the experimental cycles and technical differences between these three techniques:
Isotope in situ hybridization (RISH)
Experimental Period:
The experimental cycle of RISH is usually long because it involves the use of radioisotopes, including the preparation of labeled probes, hybridization, washing, exposure, etc. Exposure times can take anywhere from a few days to a few weeks, depending on the radioisotope used and the intensity of the signal.
Technical features:
- Probes labeled with radioisotopes such as ^32P or ^35S;
- High sensitivity and specificity, suitable for the detection of low-abundance mRNA;
- special equipment and safety measures are required for the handling of radioactive materials;
- Results are usually visualized by autoradiography and are not suitable for multiplexing;
- Number of indicators: RISH can usually only detect one indicator at a time, i.e., a specific nucleic acid sequence. This is because radiolabeled probes can only bind to a specific complementary sequence, and the detection of radioactive signals is often not suitable for multiplexing.
Digoxin in situ hybridization (DIG-ISH)
Experimental Period:
The experimental cycle of DIG-ISH is relatively short, and can usually be completed within a few days. Steps include probe labeling, hybridization, washing, enzyme-labeled antibody detection, and color development.
Technical features:
- Use digoxigenin (DIG)-labeled probes, which are non-radioactive markers;
- Signal amplification and detection using anti-DIG antibodies and enzymes such as peroxidase or alkaline phosphatase, using the principle of enzyme-linked immunosorbent assay (ELISA);
- Suitable for nucleic acid detection of tissue sections and cells;
- Results are visualized by chromogenic substrates, which are not suitable for multiplexing;
- Number of metrics detected: DIG-ISH is also largely limited to one metric at a time. While different sequences can be detected by altering the probes and corresponding antibodies, it is uncommon to detect multiple indicators simultaneously in a single experiment.
Fluorescence in situ hybridization (FISH)
Experimental Period:
The experimental cycle of FISH is also relatively short, usually completed within a few days. Steps include labeling of the probe, hybridization, washing, and detection of the fluorescent signal.
Technical features:
- Use fluorescently labeled probes such as fluorescein Cy3, Cy5; TSA dyes, etc.;
- High sensitivity and multiplexing capabilities, which can detect multiple nucleic acid sequences in the same sample;
- Results are visualized by fluorescence microscopy, suitable for quantitative and image analysis;
- Overlapping of photobleaching and fluorescence signals needs to be avoided;
- Number of test indicators: FISH technology has obvious advantages in multiplexing. By using fluorescent labels of different colors, multiple nucleic acid sequences can be detected in a single experiment. For example, fluorescent probes of different colors can be used to label different chromosomes or genes, allowing multiple indicators to be detected simultaneously. Theoretically, the number of indicators that can be detected by FISH is limited only by the detection capacity of the fluorescence microscope and the number of fluorescence channels available. Modern multiplex FISH techniques, such as spectral FISH and multicolor FISH, can detect up to dozens of different nucleic acid sequences simultaneously.
summary
- Sensitivity: RISH typically has the highest sensitivity and is suitable for the detection of low-abundance targets;
- Multiplex detection: FISH is best suited for multiplex detection, which can detect multiple nucleic acid sequences at the same time;
- Safety: Both DIG-ISH and FISH are non-radioactive, safer to operate, and do not require special facilities for the disposal of radioactive materials;
- Visualization: Results of FISH can be visualized directly by fluorescence microscopy, while RISH and DIG-ISH require additional development or chromogenic steps.
When choosing a technique, you need to consider the specific needs of your experiment, including factors such as the number of metrics to be tested, the type of sample, the cost of the experiment, and the equipment available.
【Xiaoai New Product Express】
In situ hybridization has been widely used to detect the location and number of target proteins or DNA molecules on cell or tissue sections, but for the detection of most target RNA molecules, traditional methods often cannot obtain good detection results due to the lack of specificity and sensitivity.
The GeneScope Multicolor Fluorescent Nucleic Acid in Situ Assay Kit addresses both specificity and sensitivity by utilizing an innovative combination of short probes and a unique stem-loop structure nucleic acid protein cascade signal amplification system to detect as little as 1 copy of the nucleic acid molecule of interest on the sample in situ (as shown below). Each nucleic acid to be tested will be labeled with horseradish peroxidase in the reaction, followed by a specific fluorescent substrate, and then labeled with a fluorescent molecule of that specific wavelength in a diamine-catalyzed reaction before proceeding to the next round of labeling. After up to three rounds of reaction, three channels of fluorescence can be labeled separately to detect three nucleic acid molecules. The whole experiment takes about 6-10 hours.
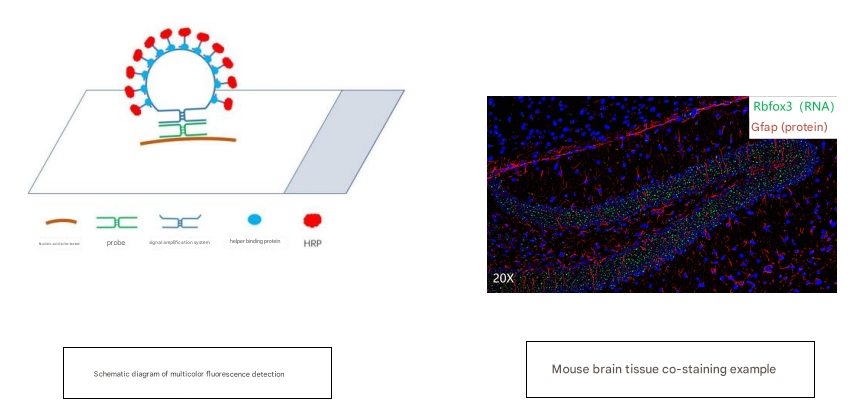
For more product information, please contact https://www.absin.cn/FISH%20Kit.html
|
Catalog number |
name |
specification |
|
GeneScope Multicolor Fluorescent RNA in Situ Assay Kit (Single) |
40T |
|
|
GeneScope Multicolor Fluorescent RNA in Situ Assay Kit (Dual Color) |
40T |
|
|
GeneScope Multicolor Fluorescent RNA in Situ Assay Kit (3 colors) |
40T |
|
|
GeneScope Multicolor Fluorescent DNA in Situ Assay Kit (Monochrome) |
40T |
|
|
GeneScope Multicolor Fluorescent DNA in Situ Assay Kit (Dual Color) |
40T |
|
|
GeneScope Multicolor Fluorescent DNA in Situ Assay Kit (3 colors) |
40T |
|
|
GeneScope Cellular RNA Fluorescence in Situ Assay Kit (Single) |
24T |
|
|
GeneScope Cellular RNA Fluorescence in Situ Assay Kit (Dual Color) |
24T |
|
|
GeneScope Cellular RNA Fluorescence in Situ Assay Kit (3 colors) |
24T |
|
|
GeneScope Cellular RNA Fluorescence in Situ Assay Kit (4 colors) |
24T |
Absin provides antibodies, proteins, ELISA kits, cell culture, detection kits, and other research reagents. If you have any product needs, please contact us.
|
Absin Bioscience Inc. |
 Follow us on Facebook: Absin Bio Follow us on Facebook: Absin Bio |
July 01, 2025
Clicks:101
